In CCMI 2.0, we will perform network mapping on breast cancer, head and neck cancer, and lung cancers, focusing on the PI3K/AKT/mTOR axis and TP53 signaling.
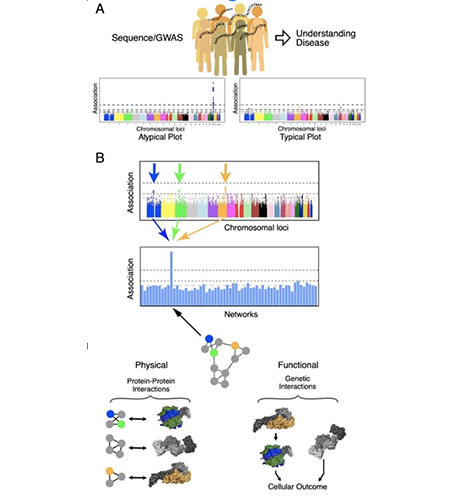
A. Current paradigm for genome wide association studies (GWAS) seeking to associate genetic variation in single genes with incidence of disease or disease outcome. With all genes tested individually for association few are typically able to pass a genome-wide level of significance. B. Mutations in genes collectively belonging to a protein assembly to predict disease outcome. Multiple genetic variants, each of which is observed rarely in a disease population, alter the activity of a protein assembly. Any mutation in this protein assembly predicts disease or disease outcome.
Integration of protein interaction data to identify protein assemblies (left). Hierarchy of protein assemblies under selection in cancer (center). Validation with clinical data and functional experiments (right).