
The purpose of this FAQ is to provide a quick reference guide for researchers involved in any Cell Maps Initiative thus facilitating access to data, interaction and collaboration within the Network Data Exchange (NDEx) infrastructure. This FAQ can also be used as a reference by anyone interested in creating a collaborative project focused on generating networks from different data sources.
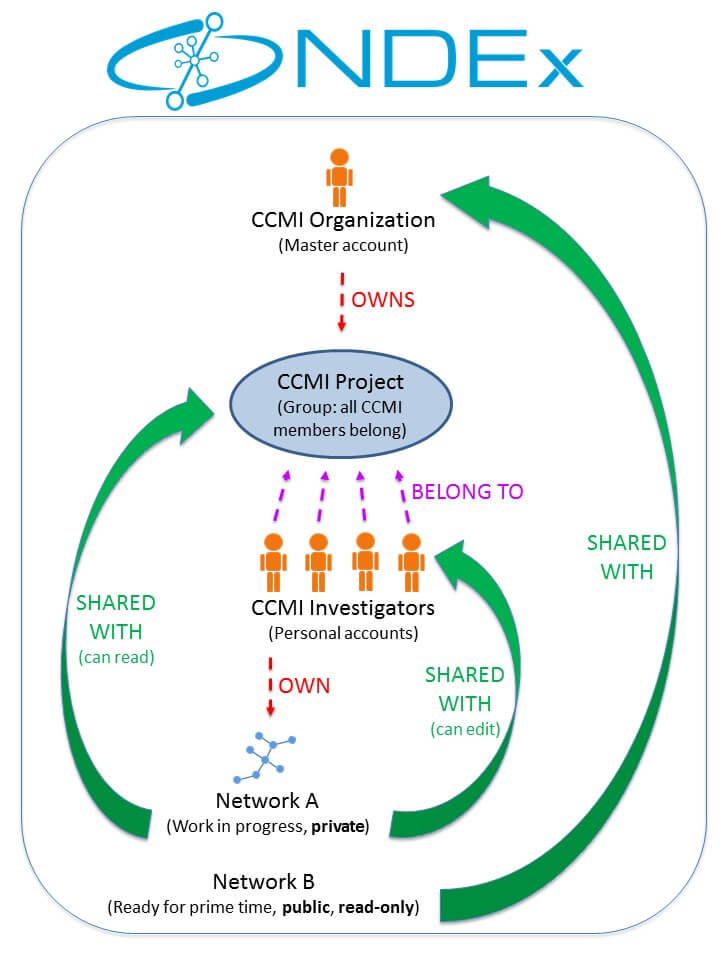
This figure below is a schematic representation of the Cancer Cell Maps Initiative (CCMI) architecture and workflow within NDEx. If you have questions or would like to recommend additions/changes to this page, please Contact Us and the NDEx Team will be happy to help!
Currently, there are 2 main ways to load a network to your NDEx account as summarized in THIS PAGE:
We have several Python example in the form of Jupyter Notebooks; JavaScript and R examples are available in THIS TUTORIAL instead.
First of all, load your networks in cytoscape using the CyNDEx App and make them look good by applying your favorite layout and visual style. Once your networks are saved back NDEx (again using the CyNDEx App), edit the network "profiles" and "properties" in NDEx to add:
Select the network you want to share and click the “Share” button. You can then search for users or groups to share your network with and decide the level of access they are allowed (Read, Edit or Admin for networks and Member or Admin for groups). When done, click the green “Save Changes” button. You can also gain access to a network you are interested in by requesting permission to its Owner/Admin. Simply select the network you would like to gain access to and click the “Ask for Access” button. For more details, review THIS PAGE.
We take security and privacy very seriously; the NDEx Public Server is hosted by Amazon Web Services (AWS), the database is backed up daily to minimize the detrimental effects in the remote eventuality of data loss/corruption, and you always control who has access to your networks:
You can create as many workgroups as you want and invite other NDEx users to join them for collaboration.
Yes, you can do this thanks to Cytoscape's built-in CyNDEx-2 core component. Please review THIS PAGE for a detailed guide and more info.
Yes, you decide who has permission to edit your networks and can always revoke permissions if you want to.
Once your network is published in a peer-reviewed journal or is ready to be disseminated to the public, the only things left to do are:
That is up to you! However, make sure you check your publisher’s policy concerning the dissemination of data pre-publication.