The NeST (Nested Systems in Tumors) map is a hierarchy of 395 protein systems that are recurrently mutated in one or more cancer types. It provides a resource of cancer mechanisms under selection for somatic mutations and captures novel protein assemblies on which mutations unexpectedly converge.
Read moreThe DNA damage response (DDR) ensures error-free DNA replication and transcription and is disrupted in numerous diseases. An ongoing challenge is to determine the proteins orchestrating DDR and their organization into complexes, including constitutive interactions and those induced by genomic insults.
Read moreAffinity purification combined with mass spectrometry (AP-MS) is used to catalog protein-protein interactions (PPIs) for 40 proteins significantly altered in breast cancer, including multidimensional measurements across mutant and normal protein isoforms and across cancerous and noncancerous cellular contexts.
This map uncovers 771 interactions from cancer and noncancerous cell states, including WT and mutant protein isoforms and enhances our understanding of head and neck squamous cell carcinoma.
Read moreMulti-Scale Integrated Cell (MuSIC) is a hierarchical map of the eukaryotic cell architecture created from integrating immunofluorescence images in the Human Protein Atlas with affinity purification experiments from the BioPlex resource.
Read moreDrugCell is a “visible” neural network (VNN) that predicts anti-cancer drug responses by modeling the hierarchical organization of a human cancer cell. Genotypes and drug structures induce differential patterns of activity on cellular subsystems, enabling in silico investigations of the molecular mechanisms underlying cancer drug response.
Read moreDCell is a deep neural network model of budding yeast, a basic eukaryotic cell. The model structure corresponds exactly to a hierarchy of 2,526 cellular subsystems. Given this neural network structure, DCell has been trained to translate genotype to phenotype.
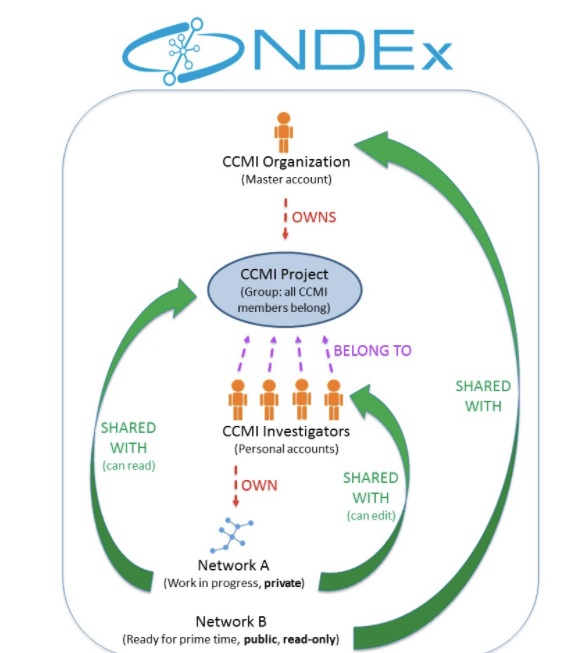
Read moreAll cell maps generated by the CCMI will be distributed via the Network Data Exchange (NDEx) Project.
To learn more about NDEx, please visit the Technology page.